Context
Hosted by the University of Washington, DubHacks is one of the largest reoccurring northwest collegiate Hackathons of the year. This time the theme was centered on creating new, and innovative technologies that would help benefit Health, Social Justice, Accessibility, and Education.Role: Project Manager, Front-End Developer, Molecular Biologist, and Sole Designer/UX Researcher.

Live Video Demo
Technologies Incorporated: This was a web application created predominantly in the M.E.A.N Software Stack (MongoDB, Express, Angular, Node.JS), with a hint of D3.js, and Particles.Io.
Conclusion: Genesis saves researchers time by handling all the Nitti gritty computational details via a simplified GUI. No more searching up individual genomic strands for cross comparisons. No more dealing with overly complicated archaic API’s. No more hand calculations to guess and check family lineages, and best of all it’s less prone to human error.
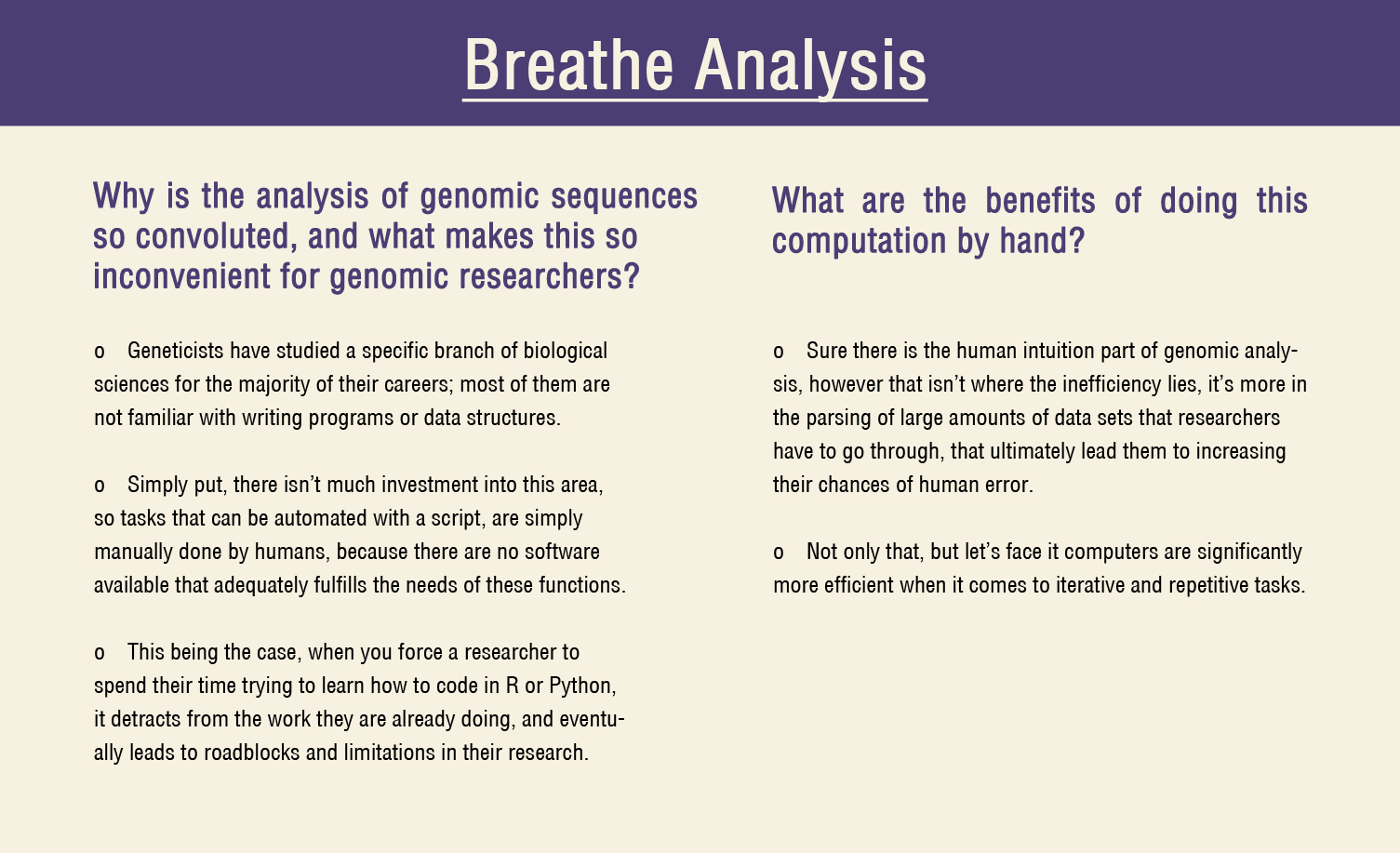
Problem
- Researchers have to do a lot of hem and hawing from different sources to retrieve data and import them to the appropriate programs for parsing.
- There is also a lot of intuition, and hand calculations that have to be done manually in order for them to get a rudimentary understanding of their target genome.
- There are no easy to use, free, and or non-programmer friendly tools for generating digitized phylogenetic lineages.
- The lack of visualizations makes interpreting data an equally cumbersome task.
*This being the case, the frequency at which researchers can analyze genomes is exponentially slower when compared to the computational power of a super computer. Even after computing the data, it takes additional effort to interpret it.
[Breathe Understanding of the Problem.]
Stakeholder Interviews (4 Total)
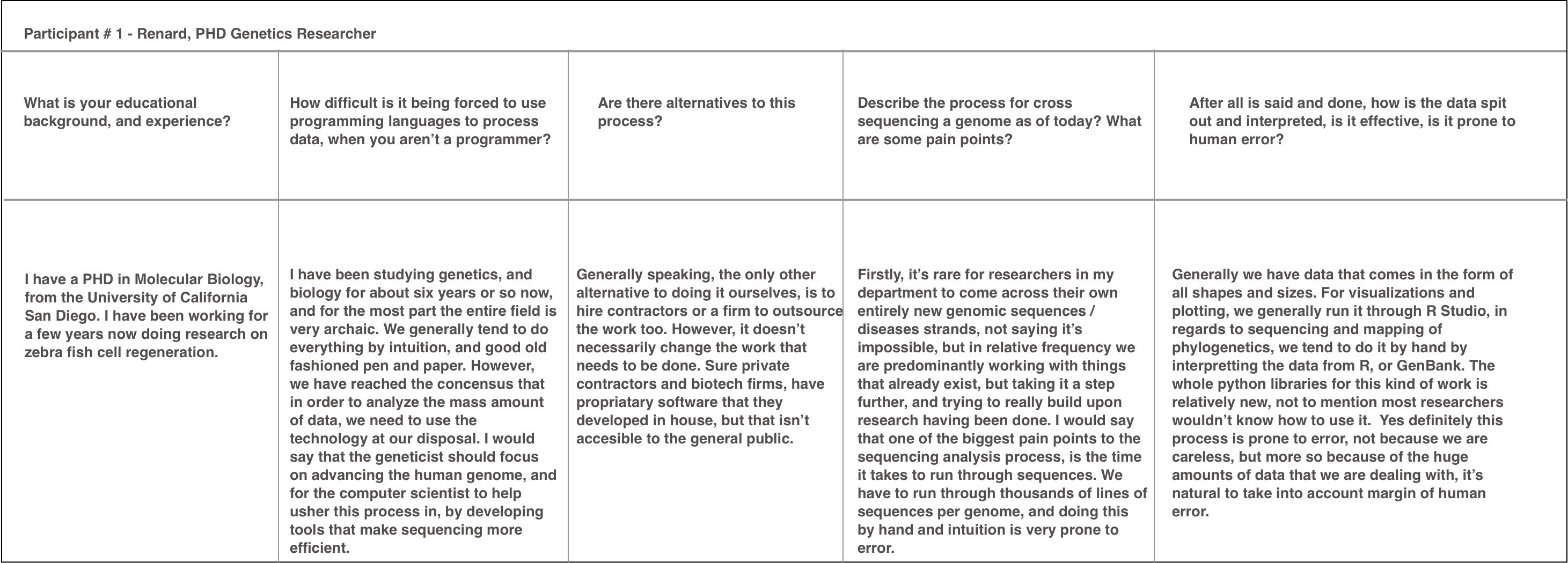
[Sample Contextual Interview Notes To Uncover Pain Points.]
*Due to the time sensitivity of Hackathons, I was limited to 4 in person interviews. Luckily however, I was able to reach out to 3 PHD researchers, and a recent graduate from the University of Washington.
Visualizing The Problem
I combined my understanding of the genomic sciences with the information gathered from the above interviews to map out the information flow. This allowed us to better analyze the redundancies within the current system.
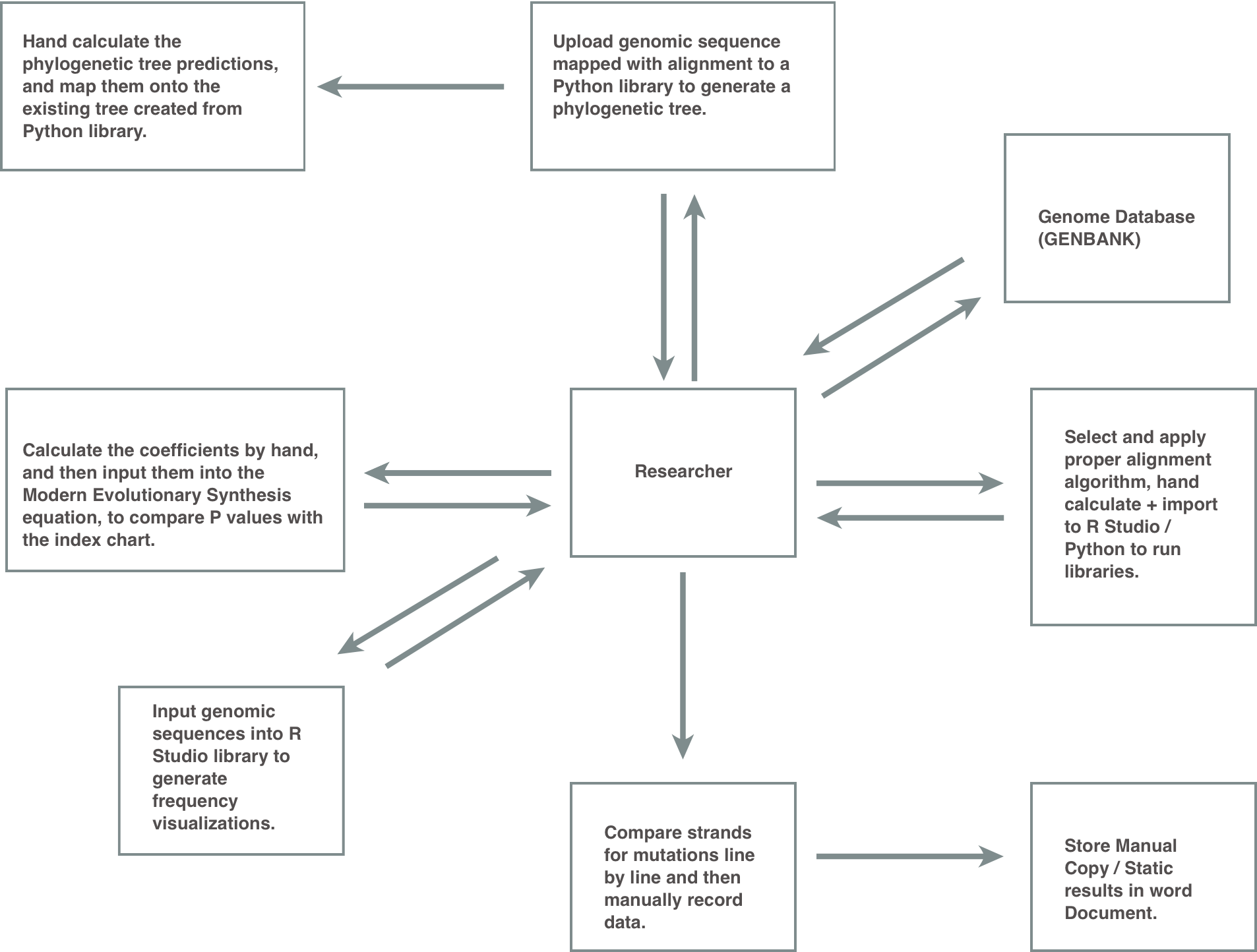
[How To Analyze A Genome Sequence.]

[My Genome 404 Class Notes.]
Hypothesis
Creating a genomic analysis tool, that compiles the previous segregated tasks into one easy to use multi-click platform, will allow researchers to get the data they want more efficiently, and with less susceptibility to human error.
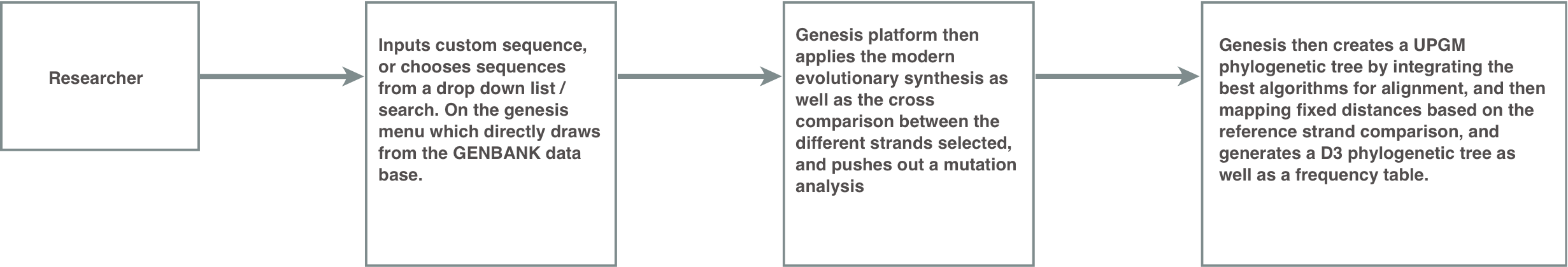
[Information Flow Following Creation Of Tool.]
User + Website Flows
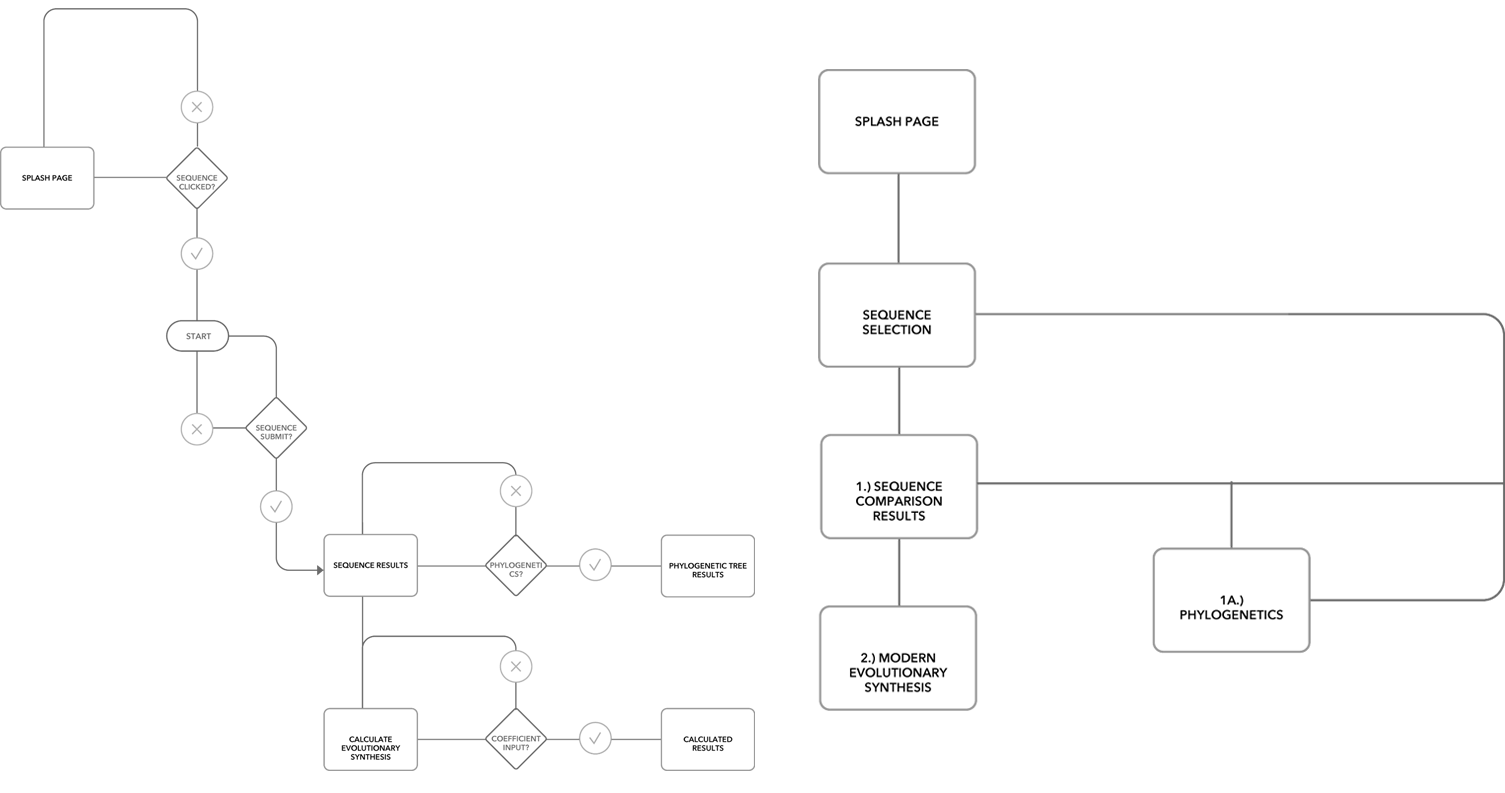
Wireframing

[Second Iteration Wireframes.]
Presentation Layout

[Splash Page with Particles.Js.]

[Gene Sequencing Comparison Page (Inspired by Diva Limous).]
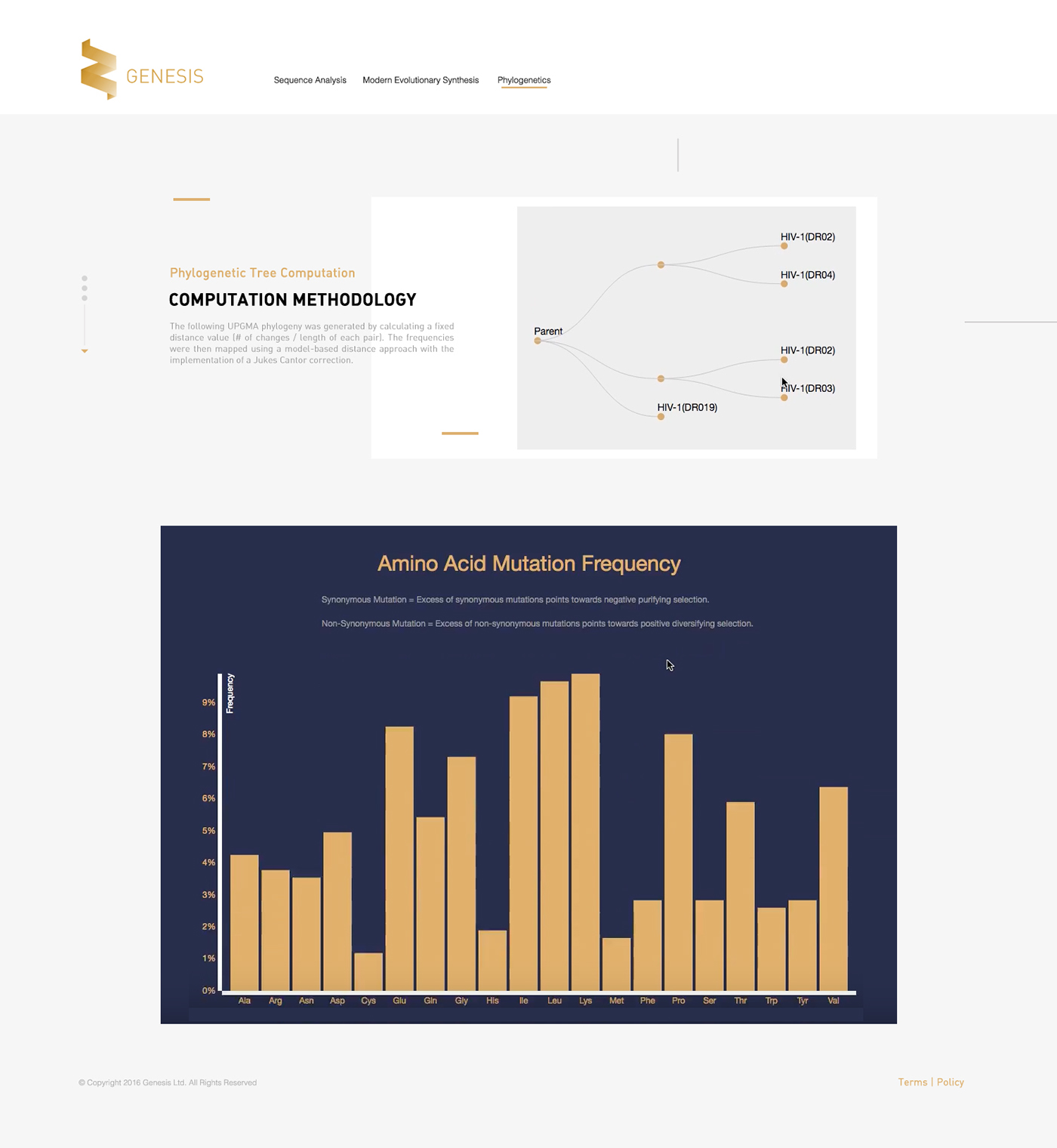
[Gene Phylogenetics Mapping Page.]


